 Click here for PDF Click here for PDF
Turmeric Raw Material and Products Laboratory Guidance Document
By John H. Cardellina II, PhDa
aReevesGroup,
Virginia Beach, VA 23451
Corresponding
author: email
Keywords: turmeric, adulteration, Curcuma
longa, Curcuma domestica, Curcuma
zedoaria, Curcuma aromatica, Curcuma
zanthorrhiza, Curcuma malabarica,
curcuminoids, synthetic curcumins, organic colorants, inorganic colorants,
analytical methods
Citation (JAMA style):
Cardellina II JH. Turmeric raw material and products laboratory guidance
document. Austin, TX: ABC-AHP-NCNPR Botanical Adulterants Prevention Program.
2020. CONTENTS 1. Purpose 2. Scope 3. Common and Scientific Names 3.1 Common name 3.2 Other common names 3.3 Accepted Latin binomial 3.4 Synonyms 3.5 Botanical family 4. Botanical Description 5. Identification and Distinction of Turmeric rhizome Using Macroanatomical Characteristics 6. Identification and Distinction of Turmeric Rhizome Using Microanatomical Characteristics 7. Genetic Identification and Distinction 8. Chemical Identification and Distinction 8.1 Chemistry of C. longa Figure 1: Representatives of the main classes of secondary metabolites in turmeric
(main curcuminoids, 1-3; prominent components of the essential oil, 4-8) and
common chemical adulterants (9, 10)
8.2 Chemistry of adulterants 8.2.1 Pigments, dyes, colorants 8.2.2 Synthetic curcuminoids 8.2.3 Other species of Curcuma 8.3 Laboratory Methods 8.4 Comments 8.4.1 Inorganic colorants Table 1: Summary comparison of approaches to determine inorganic
colorants/adulterants in turmeric raw material/commercial products
8.4.2 Synthetic organic colorants Table 2: Summary comparison of different approaches to determine synthetic
organic adulterants in turmeric raw material/commercial products
8.4.3 Curcumin content and species verification Table 3: Summary comparison of TLC, UV/VIS, and NMR methods to determine
curcuminoid content in turmeric raw material/commercial products
Table 4: Summary comparison of HPLC methods to determine curcuminoid content in
turmeric raw material/commercial products
8.4.4 DNA analysis for species verification Table 5: Summary comparison of DNA methods to verify Curcuma species identification and detect and identify adulterant
species
8.4.5 Detection of synthetic curcumin by 14C isotope measurements 9. Conclusions 10. References
1. Purpose
Turmeric (Curcuma longa
L.) dietary supplements, including standardized or partially purified extracts
with high concentrations of curcumin, have enjoyed sustained sales growth in
the United States over the past 5-6 years,1,2 while turmeric powder
continues to be an important spice, flavor, and colorant in many regions of the
world. There is considerable evidence that both powdered root and rhizome, as
well as root and rhizome extracts, have been subjected to adulteration.2 This document should be viewed in relation to the corresponding Botanical
Adulterants Prevention Bulletin on turmeric published by the ABC-AHP-NCNPR
Botanical Adulterants Prevention Program.2
2. Scope
The sustained sales growth
of turmeric supplements in the marketplace has resulted in a supply-demand
cycle that appears to have triggered considerable economically motivated
adulteration with a variety of natural and synthetic colorants and/or admixing
with other species of Curcuma. More
recently, synthetic curcumin* has been detected in some putative turmeric
products. Different analytical methods of varying complexity and expense are
required to detect and identify adulterating colorants, synthetic curcumin, and
other species of Curcuma. This document
will summarize and discuss those methods that appear most adaptable and
effective to detect these forms of adulteration.
The evaluation of a
specific analytical method or methods in this Laboratory Guidance Document for
testing turmeric materials does not reduce or remove the responsibility of
laboratory personnel to demonstrate adequate method performance in their own
laboratories using accepted protocols outlined in various domestic or
international legal and/or regulatory documents. Such documents include, for
example, the 21 CFR Part 111 (Dietary Supplement GMPs, in the US Code of
Federal Regulations) and Part 117 (Food Safety and Modernization Act [FSMA]
Final Rulemaking for Current Good Manufacturing Practice and Hazard Analysis
and Risk-Based Preventive Controls for Human Food, in the US Code of Federal
Regulations), and by AOAC International, International Standards Organization
(ISO), World Health Organization (WHO), and the International Council on
Harmonisation (ICH).
3. Common and Scientific Names
3.1
Common name: turmeric
3.2
Other common names:
Arabic:
kurkum3,4
Assamese: halodhi3,5,6
Bengali: holud (হলুদ)3,5-7†
Burmese:
tanum5
English: common turmeric, curcuma,8,9
yellow ginger5,6,9
Cambodian:
ro miet10
Chinese:
jiang huang (姜黄), huang si yu jin (黄丝郁金),9,11,12‡
jianghuang13
Danish: gurkemeje4,13
Dutch: geelwortel3,4
Filipino
(Tagalog): dilau, luyang-dilau14
French: curcuma, safran des Indes13
German: Kurkuma, Gelbwurz13
Hindi: haldi, haldee (हल्दी)6,13,14
Italian: curcuma, zafferano delle Indie3,4
Japanese: ukon3,4
Laotian: khi min10
Malay: manjal, maññaḷ (മഞ്ഞൾ)†
Marathi: halad (हळद)3,5,7
Nepali: besar,15 besaar (बेसार)†
Norwegian: gurkemeie7
Portuguese: açafrão-da-Índia3,4
Russian: yellow
ginger – жёлтый имбирь (zholtyj
imbir), curcuma – куркума3,15
Spanish: curcuma3
Sanskrit:
haridra6,8
Swedish: gurkmeja3,5,7
Tamil: manjal (மஞ்சள்)3,5-7
Telugu: pasupu (పసుపు)3†
Urdu: haldi (ہلدی)5-7,16
Vietnamese: nghệ, uất kim9,15
3.3 Accepted Latin binomial:
Curcuma longa L.
3.4 Synonyms: Curcuma domestica9,13,17,18
3.5 Botanical family: Zingiberaceae
4. Botanical Description
Curcuma
longa is an herbaceous
perennial that grows to 1.5 m tall. The part of the plant used is the rhizome,
which has a golden yellow color inside.3,10,19 The rhizome is used
as a fresh root, dried powder, herbal tea or, after extraction, as oleoresin,
dry extract, or tincture with 70% ethanol.9,20 The deep
orange-yellow powder known as turmeric is prepared from peeled, boiled, and
dried rhizomes of the plant.21
5. Identification and Distinction of
Turmeric Rhizome Using Macroanatomical Characteristics
Depending on its origin
and the soil conditions where it is grown, turmeric rhizomes can assume a
stout, short, cylindrical, or ellipsoidal structure, branching and generally
subterranean, and naturally contain 2–9% curcuminoids.22 The main rhizome is pear-shaped (ovate), typically up to 4
cm long and 3 cm thick. The upper part is encircled by leaf-scars; the lower
part is marked by scars of the secondary rhizomes and roots. Secondary rhizomes
are 0.5-1.5 cm thick, elongated, indistinctly ringed, and sparsely branched.23
Morphological characteristics of C. longa,
C. aromatica, C. zedoaria, and seven additional Curcuma spp. have been described and compared.24,25
Rhizomes of C. longa are generally
smaller (2-5 cm long) than those of C. aromatica
(3-5 cm long), C. zanthorrhiza (10 cm
or longer), or C. zedoaria (7-9 cm long),
and have a deep orange color compared to the yellow color of C. aromatica and C. zedoaria rhizomes. It is not clear if these features allow for
an unambiguous distinction among the species in practice.
6. Identification and Distinction of
Turmeric Rhizome Using Microanatomical Characteristics
A detailed description of
the microanatomical characteristics of C.
longa, including line drawings and color microscopic images, has been published.26
Additional publications contain drawings of microscopical features of C. longa and C. zanthorrhiza27 and microscopic descriptions of C. longa, C. aromatica, and C. zedoaria.25,
28-30 A substantial amount of commercially available turmeric is boiled
prior to drying, which gelatinizes the starch content. This certainly impacts
microanatomical features and, to a lesser extent, macroanatomical appearance
(color changes, spotting). A recent publication noted that microscopic
distinction among turmeric and its potential adulterating species, C. aromatica, C. zanthorrhiza, and C.
zedoaria is challenging, since some of the microscopic characteristics,
such as starch grains and oleoresin cells, are destroyed by boiling and the
cell structures of each species are similar.31 Microscopy is the
method of choice to detect admixture of undeclared starch, e.g., corn (Zea
mays, Poaceae), wheat (Triticum aestivum, Poaceae), rice (Oryza
sativa, Poaceae), tapioca (Manihot esculenta, Euphorbiaceae), to
turmeric powder. Starches can be detected using the iodine stain, or a xylene
mount with full polarization and sometimes with partial polarization to
highlight the size, shape and the “Maltese cross” of the various starches.
(Karen L. Henry, McCormick & Co., Inc., email to S. Gafner, November 1,
2019.)
7. Genetic Identification and Distinction
As
is the case with other botanicals that have a significant role in the food
market (e.g., pomegranate and cranberry), most genetic analyses to date have
been performed on C. longa with a view toward breeding programs32-34
and improving yields of primary active compounds, in this case the curcuminoids.35,36
However, there have also been potentially useful investigations of the genetic
diversity in species of Curcuma, with
a view toward using those differences to verify the identity of C. longa and the presence or lack of
adulteration by other Curcuma spp.
In
one such study,37 15 economically important species of Curcuma from India (C. amada, C. aromatica, C. aeruginosa, C. caesia, C. comosa, C. decipiens, C. ecalcarata, C. haritha, C. longa, C. montana, C. malabarica, C. pseudomontana, C. raktakanta, C. sylvatica, and C.
zedoaria)§ were examined by Random Amplified Polymorphic DNA
(RAPD) and Inter Simple
Sequence Repeats (ISSR) technologies to assess their genetic diversity,
polymorphism, and relatedness. UPGMA (Unweighted Pair Group Method with
Arithmetic Mean) cluster analysis of the data revealed two clusters, one
containing only two species, differentiated at a similarity of 0.57 from all
the others. The two species in this cluster are often placed in subgenera of Curcuma. The other cluster was divided
into 6 groups; C. longa was in a
group by itself and showed a similarity of 0.64 to four of those groups and
0.62 to the remaining group.
In another study of 11
“starchy” Curcuma spp. from India (C.
aromatica, C. amada, C.
aeruginosa, C. brog, C. caesia, C. haritha, C.
leucorrhiza, C. longa, C. malabarica, C. raktakanta,
C. sylvatica, and C. zedoaria),38 RAPD was utilized to compare the DNA profiles
and UPGMA was used to develop relatedness dendrograms. As in the first study, C. longa is not closely related to any
of the other species; in this case, the closest relative was C. zedoaria, with a similarity
coefficient of 0.7, while the others were in the 0.55-0.68 range. It is
interesting to note that, although The Plant List18 regards C. brog as a synonym for C. longa,
this study found C. brog more closely related to C. aromatica and
C. leucorrhiza.
The same RAPD/UPGMA
approach was used in a third study, this time of 12 identified (C. aeruginosa, C. albicoma, C. amada, C. angustifolia, C. aromatica, C. comosa, C. longa, C. mangga, C. parviflora, C. petiolata, C. rubrobracteata, and C.
sessilis) and three unidentified Curcuma
spp. samples from Thailand.39 As seen in the dendrograms from
the other reports cited above, C. longa has
only C. zedoaria as a close relative
(0.93 similarity). Overall, the species were organized by relatedness into
three clusters; one contained only C. parviflora,
and the second was comprised of C. petiolata
and C. rubrobracteata (0.83
similarity). The remaining large cluster was subdivided into four groups; the C. longa/C. zedoaria group is related to the other three groups by a
similarity factor of 0.36. It is interesting to note that the three
unidentified Curcuma spp. were more
closely related to one another (0.76, 0.70 similarity) than to any of the other
species. It should be noted that this report lists C. albicoma as one of the species in this study in Table 1 of the
article, but all subsequent discussion of the results provides no mention of C. albicoma, but does provide results
and discussion on C. zedoaria instead;
a possible explanation is that the taxonomy was revised from C. albicoma to C. zedoaria late in the
study, and the name was simply not corrected everywhere in the manuscript.
Even though the same
technologies were used in these three studies, there are some distinct
differences among the results of the three studies; further, the taxonomic inconsistencies
in both studies might raise questions about the security of those
identifications. However, the one factor that does stand out is that C. longa does not have many, if any,
close genetic relatives in the genus. This suggests that genetic testing can
and likely will be a useful tool for identification of fresh and dried C. longa in commerce.
Yet another genetic method40
was proposed to distinguish among turmeric and its potential adulterating
species, C. aromatica, C. zanthorrhiza, and C. zedoaria via chloroplast DNA
polymorphisms in the trnS-trnfM intergenic
spacer region; all four species were correctly identified. Further,
curcumin content in C. longa rhizomes could be predicted by the number of AT repeats in the trnSfM region.
More research is needed to
determine how certain processing steps (e.g., heating, extraction, filtration)
affect the ability of methods based on DNA markers to identify ingredients made
from turmeric. Obviously, the DNA-based methods will not provide information
about the plant part(s) present, adulteration with undeclared dyes, or detect
the addition of synthetic curcuminoids to turmeric extracts.
8. Chemical Identification and Distinction
8.1
Chemistry of C. longa
While there continues to
be disagreement and debate about the taxonomy of the genus Curcuma, there are at least 100 species, but only 20 of them have
been the subject of any significant investigation of their chemistry.41
Curcuma longa is by far the most
thoroughly studied species of the genus; a 2011 review showed that 235
different secondary metabolites had been reported from C. longa to that point in time.21 Several structural
classes of natural products are found in C.
longa, including diarylheptanoids (curcuminoids), the structurally related
diarylpentanoids, a large number of monoterpenes and sesquiterpenes – primarily
responsible for the aroma and flavor of turmeric, and significantly smaller
numbers of diterpenes, triterpenes, and sterols (see Figure 1). A few, rather
common fatty acids have also been identified in the rhizomes. The
diarylheptanoids are important for their relative abundance, color, and purported
pharmacological activity. The three most abundant curcuminoids in C. longa are curcumin (1), demethoxycurcumin (2), and bisdemethoxycurcumin (3).
Monoterpenes and sesquiterpenes are quite numerous in the
volatile (essential) oil of turmeric. As of the writing of the 2011 review
cited above,21 69 monoterpenes and 106 sesquiterpenes had been
reported from C. longa. Monoterpenes
dominate the essential oil of the leaves and flowers (aerial parts), while sesquiterpenes
comprise the majority of the rhizome essential oil. A simple test for
adulteration of rhizome material with aerial parts would be the presence of
significant amounts of monoterpenes. Some representative sesquiterpenes from C. longa (see Figure 1) include ar-turmerone
(4), α-turmerone (5), and β-turmerone (6).43 These
compounds may account for 40% or more of the essential oil of turmeric rhizomes.44
Additional sesquiterpenes first found in C.
longa include turmeronols (e.g., 7)45
and curculonones (e.g., 8).46
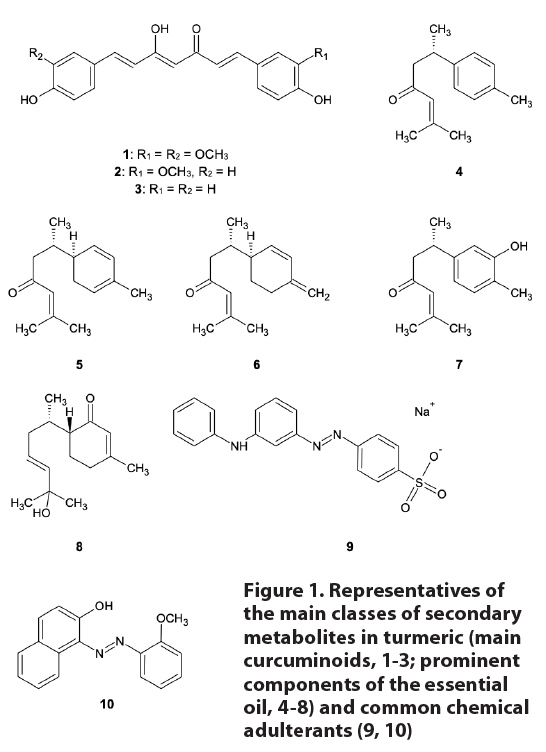
8.2
Chemistry of adulterants
The adulterants of
turmeric (C. longa) include inorganic
and synthetic organic dyes/pigments to adjust the color of the raw material,
exogenous curcumin, and other species of Curcuma.
Figure 1 also provides the chemical structures of some of the most important
coloring agents that have been found as adulterants in turmeric.
8.2.1
Pigments, dyes, colorants
Lead chromate, PbCrO4,
is a somewhat rare mineral found in the oxidation zones of lead ore beds.
Synthetic lead chromate is a bright yellow inorganic pigment used in paints. It
is inexpensive and easy to prepare, and readily available; unfortunately, it
has far too often been detected bolstering the color of substandard or
fraudulent turmeric (and other spice) products.46-51 Since both lead
and chromium are among the heavy metals of greatest health safety concern, the
use of this compound to color fraudulent or diluted turmeric samples goes
beyond just intentional economic adulteration to a potentially serious public
health safety issue.
Several synthetic organic
dyes or pigments have also been used to enhance or add color to supposedly
authentic turmeric samples. The sodium salt of Metanil Yellow (9) is used as a pH indicator, but it
has not been approved as a food additive or food ingredient. It has nonetheless
been found as an adulterant in turmeric.52,53 Sudan Red G (10) is another azo dye once used as a
coloring agent for fats and waxes; it was formerly used as a food coloring
agent, but is now banned for that use, as the European Food Safety Authority
considers it genotoxic and/or carcinogenic.54 It, too, has been
reported as an adulterant in turmeric.55
8.2.2
Synthetic curcuminoids
Synthetic curcuminoids can
be most effectively distinguished from their naturally biosynthesized counterparts
by evaluation of the amount of 14C found
in a sample under investigation. Natural products, in this case curcuminoids,
are prepared from CO2 by photosynthesis, incorporating a consistent
level of 14C into each compound. Synthetic curcuminoids are typically
prepared from petroleum-derived chemical feedstocks (starting materials) and
have exceedingly low-to-no detectable 14C present. Furthermore,
curcumin (1) and bisdemethoxycurcumin (3) are easier and cheaper
to synthesize, since the phenylpropane moieties of those molecules are
identical.
The three major
curcuminoids of C. longa, curcumin, demethoxycurcumin, and
bisdemethoxycurcumin, are typically found in partially purified extracts of C. longa in ratios ranging from 20:9:5
to 7:2:1.56 Although this ratio is primarily dependent on the
cultivar, it may also be affected by storage conditions and
extraction/purification protocols. Any significant deviation from this ratio
could suggest adulteration with synthetic curcumins, if all three major
compounds were not prepared and mixed in the proper ratio. Alternatively,
adulteration by, or substitution with, other species of Curcuma with different curcuminoid content might be the
explanation.
8.2.3
Other species of Curcuma
Curcuma
longa has been
reported to be adulterated with C. zedoaria,57-62 C. aromatica,57 C. zanthorrhiza,63 and C. malabarica.64 While C. zedoaria would seem to be the most
common adulterant of C. longa, there
are insufficient reports of the testing of numbers of commercial samples of purported
C. longa to support that assignation.
On the other hand, it is interesting to note that multiple DNA studies of
numerous species of Curcuma (see
Section 7, above) found that C. longa
is most closely related to C. zedoaria and distinctly different from
all other species tested. Further, C.
zedoaria and C. aromatica also contain curcuminoids, making C. zedoaria a potentially
attractive candidate to replace or dilute C.
longa in the supply chain.
Curcuma
aromatica: The three major
curcuminoids (1-3) in C. longa
are also found in C. aromatica,
albeit in lower concentrations. One report indicated that total curcuminoids
vary between 0.03-0.3% in C. aromatica,65
while another reported up to 1.3% curcumin.66 Curcumin is the
dominant diarylheptanoid, while demethoxycurcumin and bisdemethoxycurcumin
reportedly are present at similarly low concentrations. More recent papers
suggest that bisdemethoxycurcumin is not found in C. aromatica, and can be used as a means to detect adulteration.60,67,68
The essential oil composition varies substantially, depending on the sample,
and some of the published results may be due to inaccurate species
identification, since a review of the published data indicated that camphor (18-36%),
1,8-cineol (5.5-12%), α-curcumene (0.3-25.7%), ar-turmerone (2.5-18%),
and curzerenone (5.3-11%) are frequently present in this essential oil.69
Curcuma
zanthorrhiza: The
content in curcumin and demethoxycurcumin is between 0.8-2.0%, with a 1.7:1
ratio of the two components.65,70 Bisdemethoxycurcumin is present only
in traces, or not detected in the roots. The absence of this compound can be
used as a means to detect adulteration with C.
zanthorrhiza. The essential oil is mainly composed of sesquiterpenes;
α-curcumene (13-65%), β-curcumene (16-17%), and xanthorrhizol (20-32%) make up
the majority of the essential oil.69,70 Xanthorrhizol is considered
a marker compound for C. zanthorrhiza.70
Curcuma zedoaria: The vernacular name “white turmeric,”
sometimes used for C. zedoaria, is due
to the color of the roots, which are white or light yellow on the inside
because of low concentrations of the orange-colored curcuminoids present.67,71,72
Actual data on the concentration of these curcuminoids in dried zedoary are quite
limited; demethoxycurcumin is the predominant curcuminoid in zedoary, making up
0.003% of the dried root, about 10 times more than curcumin.71 The higher
concentration of demethoxycurcumin (relative to curcumin) can be used as an
indicator for adulteration with C. zedoaria.
Zedoary rhizome oil is mainly composed of
sesquiterpenoids (80–85%) and monoterpenoids (15–20%). The major components in
the essential oil reportedly vary, and include epicurzerene (19–47%), curzerene
(10-32%), curzerenone (22–32%), curdione (7–20%), and 1,8-cineole (12–41%).69
8.3
Laboratory Methods
There are quite a few
reports in the literature on analytical methods to identify turmeric, assess
its quality, and/or determine evidence of adulteration. Not all the reported
methods are necessarily suitable for all these purposes or all forms of turmeric
in the marketplace.
8.4
Comments
Given the number of forms
of adulteration to be addressed and the number of methods to be discussed, the
comments will be divided into groups based on the specific type of
adulteration. Within those groups, there may be subgroups based on the
analytical methodology used. A table summarizing the different analytical
methods selected will be provided for each section.
8.4.1
Inorganic colorants 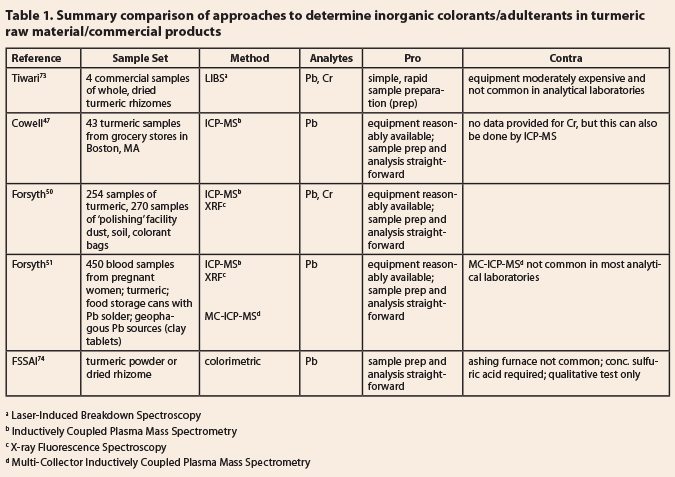 Lead chromate is a bright yellow inorganic pigment used in certain paints. Unfortunately, it
has also been utilized to color foods and spices to make them appear more
attractive or more representative of high quality. This compound provides the
double health hazard of exposure to two toxic heavy metals, lead and chromium.
Fortunately, it can be readily detected. Five methods are summarized in Table
1.
Tiwari et al.73
used Laser-Induced Breakdown Spectroscopy to establish the lead and chromium
content of four commercial samples of whole, dried turmeric rhizomes, while
Cowell et al.47 used Inductively Coupled Plasma Mass Spectrometry to
quantify the lead content of 43 commercial turmeric products obtained from
stores in Boston. Both of these techniques are characterized by simple sample
preparations, relatively quick analyses, and can be used to detect and quantify
both lead and chromium, even though Cowell’s study was focused on lead only. It
is noteworthy that one could use the lead-to-chromium ratio to gauge whether
additional lead was added to the turmeric sample in question by uptake from the
soil during growth or from water used to wash the roots post-harvest. In 2019, Forsyth
et al. have reported two studies of lead intake by the population of
Bangladesh, with an emphasis on turmeric.50,51 Both studies used
ICP-MS and XRF for the relevant analyses. The first50 focused on
turmeric samples and the dust in and soil around the facilities where the raw
turmeric was treated (polished) with lead chromate, the PbCrO4,
while the second study51 analyzed the lead content of blood drawn
from pregnant women. A unique feature of this latter study was the use of lead
isotope ratios (by MC-ICP-MS) to identify the source of the lead – turmeric,
lead solder in food storage cans, and clay used to make tablets (pills)
consumed during pregnancy. Another revelation from this study was that the
Pb:Cr ratios were all in a range of 1.2 to 1.4, rather than 1:1 as expected for
PbCrO4; the authors suggest that this is due to varying amounts of
PbCO3 and PbSO4 (lead carbonate and lead sulfate,
respectively) in the less than “reagent grade” colorant materials.
The fifth ‘method’74
is actually a compendium of various methods (colorimetric, TLC, and HPLC) for
evaluating adulteration in turmeric samples. The ashing/sulfuric acid-phenyl
carbazide test is mentioned here because it is a relatively easy, quick,
qualitative test for lead; one should follow a positive result with either
rejection of the turmeric lot or a quantitative test to confirm the presence of
lead and quantify the amount present. The ICP-MS method would seem to be the
most versatile (and available) technique available for this important and
dangerous adulterant (or contaminant; sometimes the presence of the lead is
based on accidental contamination, i.e., not intentional adulteration).
8.4.2
Synthetic organic colorants 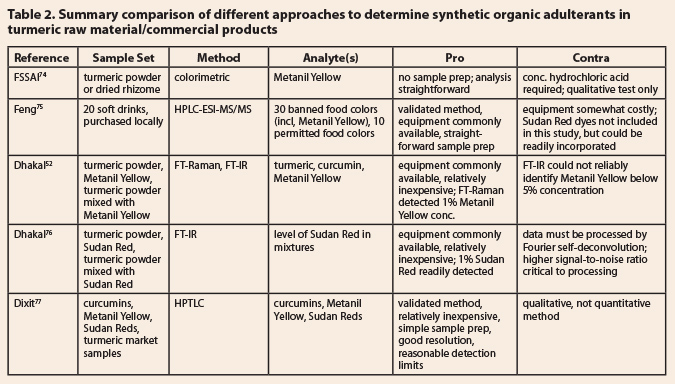 A variety of diaryl azo
dyes, including Metanil Yellow and the Sudan Dyes (see Figure 1 for examples),
have been reported as colorant adulterants in turmeric powders.52,53,55
Five analytical methods are summarized in Table 2. The simplest of these is the
validated HPTLC method of Dixit et al.77 This method is relatively
inexpensive, has a quick sample preparation, and can simultaneously determine
the presence of Metanil Yellow, the more common of the Sudan Red dyes, and the
presence of the three primary curcuminoids in turmeric. Feng et al. developed
and validated an HPLC-MS/MS method that could detect and quantify any of 30
banned colorants and 10 permitted (food) colorants.75 One drawback
to this method is that the Sudan Red Dyes were not included in the method
development, but it is likely that one could add them to the analyte pool with
minor tweaking of the HPLC method. The third method, by Dhakal et al.,52
focused on FT-IR and FT-Raman for the detection of Metanil Yellow in turmeric
powder. Unfortunately, FT-IR could not reliably detect levels of Metanil Yellow
below 5%; FT-Raman was a bit better, but the 1% detection limit still seems a
bit high for an intense colorant. However, Dhakal et al.76 were able
to develop an FT-IR method to identify Sudan Red mixed with turmeric powder at
levels as low as 1%; the extensive data processing requirement (noise
reduction, curve fitting, etc.) is a bit of a drawback to this method.
For these adulterants, the
HPTLC method77 seems to be the better choice, unless one wanted to
examine raw material or extracts for a wider variety of potential adulterant
colors. Then, the HPLC method75 would seem the logical choice. The method
by the Food Safety and Standards Authority of India (FSSAI)74 would
seem most appropriate for a qualitative test of bulk raw material samples for
Metanil Yellow. It is a simple colorimetric test, consisting of adding a few
drops of concentrated hydrochloric acid to the powder in question and observing
a pink color develop; if the color persists after dilution with water, Metanil
Yellow is present.
8.4.3
Curcumin content and species verification 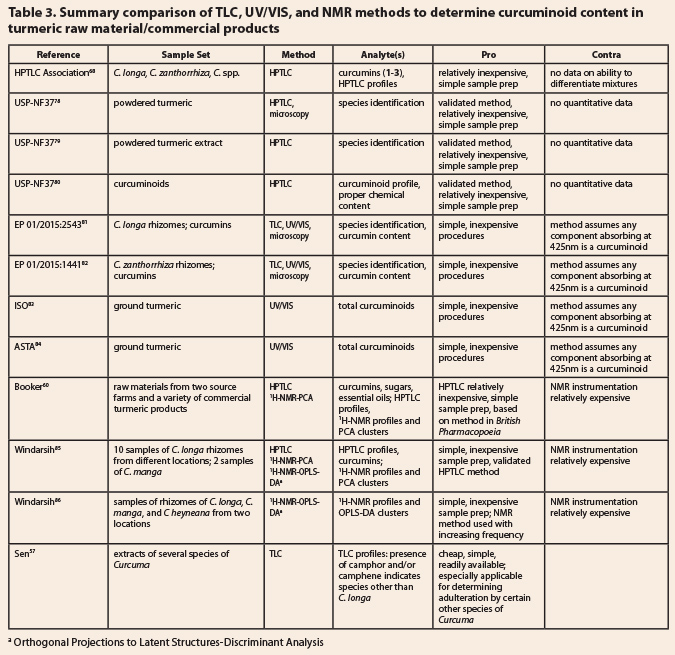 Twelve TLC, UV/VIS and NMR
methods are summarized in Table 3 and discussed here. The primary advantages of
TLC methods are speed, cost effectiveness, and the ability to detect the
presence of, or substitution with, other species of Curcuma, while the chief disadvantage is the lack of rigorous
quantitative data on the curcumin content or extent of adulteration or admixing
with other species. The HPTLC Association has developed a method to distinguish
C.
longa, C. zanthorrhiza, and an
unspecified Curcuma spp.68 This
method might be expanded to deal with other species of Curcuma, but the authors provided no data on identifying mixtures
of species. Booker et al.60 conducted a similar study, but expanded
it to include four species of Curcuma
(C.
longa, C. zanthorrhiza, C.
aromatica, C. kwangsiensis)
and both HPTLC and 1H-NMR
with principal component analyses (PCA).
Curcuma kwangsiensis is mentioned in the text, but could not be cleanly
differentiated from the other species by NMR-PCA; moreover, the sample gave
only a few faint spots on the usually information-rich derivatized TLC plates
examined under white light. HPTLC and NMR-PCA could differentiate C. longa from the other two species, but
those two species were not readily distinguished from one another by either
technique, forming a mixed cluster by NMR-PCA and giving similar HPTLC
profiles. Windarsih et al.85 have followed the report of Booker et
al.60 with a modification of the CAMAG TLC system (altered
developing solvent) and a NMR-PCA study of C. longa and C. manga,
showing that they are readily distinguished by both methods and that admixing C.
longa with varying amounts of C. manga could be detected by
conventional PCA down to 10% of C. manga. However, application of
OPLS-DA (orthogonal partial least squares-discriminant analysis) resolved all
the admixed samples from pure C. longa and C. manga. In a
contemporaneous study, Windarsih et al.86 relied solely on 1H-NMR-OPLS-DA to differentiate C.
longa, C. manga, and C. heyneana; these two studies
demonstrated that Curcuma species containing little or no curcuminoids
can be discovered when admixed with or adulterating C. longa.
It should be noted that
the HPTLC methods discussed here all employed the same method developed by
CAMAG (except for Windarsih et al.85 and Sen57) and
incorporated in the USP (United States Pharmacopeia) methods for powdered
turmeric,78 powdered turmeric extract,79 and curcuminoids.80
The CAMAG method has also been incorporated into the Ph. Eur. (European Pharmacopoeia)
monographs on C. longa81 and C zanthorrhiza.82
Additionally, the Ph. Eur. methods also provide a simple UV/VIS absorption at 425 nm to
calculate the equivalent content of curcumin in a given sample; the simplicity
and cost effectiveness of this method are dramatically offset by the ease with
which adulterants with a strong absorption at the target wavelength could
deceive an analyst. Pairing the UV/VIS test with an HPTLC analysis that shows a
solid match to a reference standard of turmeric would seem to provide a reliable
combination of methods. The ISO (International Organization for
Standardization)83 and ASTA (American Spice Trade Association)84
both utilize quite similar UV/VIS method to determine the curcumin content of
ground turmeric. As noted above, the UV/VIS method is quick and simple, but
assumes any compounds absorbing at 425 nm are curcuminoids; to rule out
adulteration, one must have a second test to rule out added colorants.
The 1974 publication by
Sen et al.57 offers a useful insight for the use of TLC; these
researchers could detect C. zedoaria
and/or C. aromatica in purported C. longa by TLC, by including camphor
and camphene as additional standards. Those two compounds are not present in C. longa, but are found in several other
common species of Curcuma. The
presence of the curcumins (1-3) in the proper ratio and the absence
of camphor and camphene in a modern HPTLC system could provide corroborating
evidence for unadulterated C. longa.
Since Dixit et al.77
used HPTLC to seek and identify synthetic colorants (Metanil Yellow and Sudan
Red), it is likely that the methods described here could be adapted to the same
purpose. None of the methods listed above can be used to differentiate natural
turmeric-derived curcumins from synthetic curcumins. 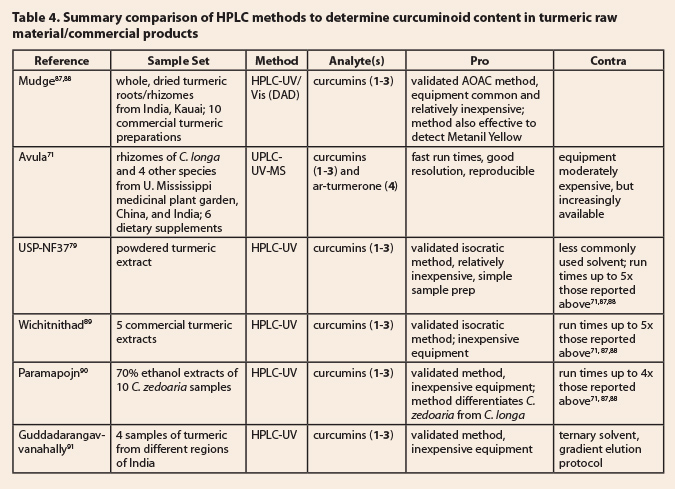 Six HPLC methods for
determining curcumin content are listed in Table 4. Mudge et al.87
reported a rapid, validated, quantitative method for the three main curcumins (1-3)
using HPLC-UV/Vis (diode array detection) and then reported this method as an
AOAC Single Laboratory Validated Method.88 Earlier, Avula et al.71 presented a rapid quantitative
method using UPLC-UV-MS to detect and quantify the curcumins (1-3)
and ar-turmerone (4), previously identified as an
anti-venom (snakebite) constituent of C.
longa.92 Both of these methods use relatively short, narrow
diameter, fine particle size columns to reduce run times and conserve solvent,
while not sacrificing resolution or peak shape. Mudge et al.87 also
established resolution and identification of Metanil Yellow in their work,
thereby providing a method that could simultaneously quantify the curcumins
present, establish the presence or absence of the adulterant/colorant Metanil
Yellow and provide evidence for adulteration by other species of Curcuma by the presence (or absence) and
ratio of the main curcumins (1-3). The approach of Avula et al.71
differed by the addition of a mass spectrometry detector, comparative analysis
of C. longa and four additional
species (C. zedoaria, C. phaecaulis, C. wenyujin and C.
kwangsiensis), and introduction of another distinguishing analyte, ar-turmerone (4), present in C. longa,
but not in any of the suspected adulterant species.
The remaining four HPLC
methods used longer, wider diameter, larger particle size columns, resulting in
longer run times (up to 5x those described above).71,87,88 The
isocratic USP method79 provides very good resolution of the
curcumins, but requires run times of nearly 30 minutes. Wichitnithad
et al.89 also developed and validated an isocratic HPLC method to
separate and quantify the curcumins of interest (1-3), but run times are
16 minutes. Commercial extracts of turmeric were used in this study; no details
of their preparation were provided. Paramapojn et al.90 used a
validated gradient elution method to examine 10 collections of C. zedoaria from different locations in
Thailand; run times are 13 minutes. The authors reported that this was the
first determination of the amounts and ratios of the curcumins (1-3) in C. zedoaria; demethoxycurcumin (2) was consistently the dominant curcumin
in this species. Earlier, Guddadarangavvanahally
et al.91 had reported a ternary elution system (methanol-2% acetic
acid-acetonitrile) to separate the curcumins of interest in about 8 minutes.
These four methods were summarized here because they represent alternative,
validated approaches to solving this separation problem.
While there are appealing aspects
in each of the six methods summarized in this section, the methods of Mudge et
al.87,88 and Avula et al.71 stand out for the validated
analytical protocols, modern column technology, shorter run times (time and
cost efficiencies), and demonstrated ability to look simultaneously for an
adulterating colorant and an additional marker relevant to the identity of C. longa.
8.4.4
DNA analysis for species verification 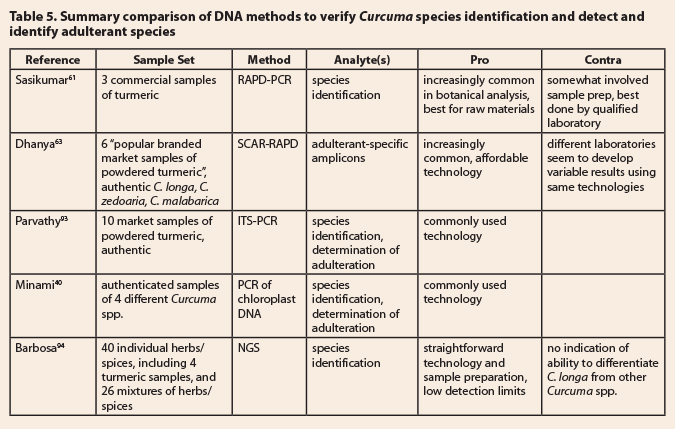 Table 5 lists three
publications from the Sasikumar group specifically focused on the detection of
adulterating species in commercial turmeric samples by DNA analyses.61,63,93
In the first study,61 the authors amplified DNA from authenticated
samples of C. longa and C. zedoaria, as well as three popular
marketplace samples of turmeric. RAPD analysis was performed using eight random
decamer primers to identify species specific markers. When this approach was
applied to the marketplace samples, the researchers found higher percentages of
C. zedoaria markers, even though the
curcumin content was in the range expected for C. longa. The second study63 expanded the scope of the
first study by adding C. malabarica as
a second potential adulterant species and by using SCAR (Sequence Characterized
Amplified Region) markers designed from two C.
zedoaria/C. malabarica-specific RAPD markers to examine genuine turmeric
and six commercial ‘turmeric’ samples. Four of the six samples were found to be
adulterated by one or both of the other species; adulteration could be detected
at levels ~1% of the total mass of the sample. In a more recent study,93
DNA barcoding was used to detect plant-based adulterants in market samples of
turmeric powder using a library of authentic rhizomes from C. longa and C. zedoaria. The genetic ITS region
contained single nucleotide polymorphisms (SNPs) specific to C. zedoaria DNA. These SNPs proved
useful in detecting adulteration; one of 10 market samples contained C.
zedoaria, one contained tapioca starch, and a third contained barley (Hordeum
vulgare, Poaceae), wheat, and rye (Secale cereale, Poaceae) flour.93
Minami et al.40 proposed an alternative genetic
method to distinguish among turmeric and its potential adulterating species, C. aromatica, C. zanthorrhiza, and C.
zedoaria, via chloroplast DNA polymorphisms in the trnS-trnfM intergenic spacer region; all four species were
correctly identified. Further, curcumin content in C. longa rhizomes
could be predicted by the number of AT repeats
in the trnSfM region. A recent report by Barbosa et al.94
is included, because the approach is a bit different and is very sensitive (low
detection threshold). The authors used NGS (Next Generation Sequencing) to
examine a large number of commercial samples, including individual herbs/spices
and mixtures. While only 4 turmeric samples were analyzed, all of them were
found to contain other herbs/spices, including fenugreek (Trigonella foenum-graecum, Fabaceae), cumin (Cuminum cyminum, Apiaceae), chili pepper (Capsicum
annuum, Solanaceae), coriander (Coriandrum sativum, Apiaceae), and
garlic (Allium sativum, Amaryllidaceae).94 Since none of
these herbs/spices have been identified as adulterants of turmeric, the logical
deduction is that the presence of these plant residues is due to poor adherence
to good manufacturing or good food practices.
While the studies cited
and discussed here are largely preliminary, in terms of dealing with the
problem of adulteration of C. longa
with other Curcuma spp., they are
indicative of considerable progress in this area. Three papers discussed earlier
in Section 7 (vide supra) may prove
to be important to developing a unified, efficient approach to genetic
differentiation of Curcuma spp.37-39
A promising aspect of this approach is that C.
longa appears to be unique in its genetic relationship to (or differences
from) other species of Curcuma, based
on all these studies, making it seemingly easier to identify as pure or
adulterated in market samples. This arena is likely to see considerable
development in the near future.
8.4.5 Detection of
synthetic curcumin by 14C isotope measurements
There are suitable
analytical methods to deal with adulterating colorants, curcumin content
(proper amount and ratio), and mixing or substitution by other species. While
there are currently no detailed scientific publications on detecting synthetic
curcumins through the use of mass spectrometry to evaluate 14C
content of the curcumins in a given sample, this technology does exist and has
been explored with regard to turmeric and curcumin origins.95 Mass
spectrometry can be employed to determine the amounts of different carbon
isotopes (12C, 13C, 14C) present in a given
sample of curcumin. True natural products have residual traces of 14C
due to photosynthesis from ambient 14CO2, whereas
curcumin synthesized from petrochemical feedstock will have no detectable 14C
content, given the short half-life of 14C relative to the age of the
petroleum source. Publications on this subject can be expected in the not-too-distant
future.
9. Conclusions
Turmeric
sales continue to grow, both in the supplement/phytomedicine and food/flavor
sectors. Thus, growing demand has put pressure on the supply chain, leading to
economic adulteration. Adulteration in turmeric can take on several forms:
- colorants
added to enhance the appearance of the raw material — these can be inorganic or
synthetic organic dyes;
- mixing
or substitution with other species of Curcuma;
- addition
of undeclared fillers, such as wheat or rice flour, to turmeric powders; and
- addition
of synthetic curcuminoids to adulterating species.
Of these forms of
adulteration, only mixing or substitution with other species might be
incidental, accidental, or unintentional, but this too can also be intentional,
especially if combined with other adulterations. The other forms of
adulteration are clearly intentional.
All raw material should be
subjected to tests for inorganic and synthetic colorants, species identity, and
curcumin content before acceptance by a manufacturer of finished products. This
may require multiple analyses. Finished products should be checked for curcumin
content, particularly if a label claim is made about that content, and
curcuminoid ratios.
Safety Issues
There are serious safety
concerns about the use of artificial colors and dyes to enhance the appearance
of substandard or false C. longa.
Lead chromate delivers not one, but two toxic heavy metals to a consumer of
adulterated turmeric. Since lead is a cumulative toxin, it represents a serious
health threat, especially to young children. A series of papers by Forsyth et
al.50,51 (and additional studies cited therein) reveal how
widespread and massive the lead exposures are in Bangladesh, including blood
levels 1-3 orders of magnitude above the maximum allowed exposure in consumers
(children, pregnant women) and workers in the shops where turmeric rhizomes are
‘polished’ with lead chromate.
The synthetic colorants,
such as Metanil Yellow and the Sudan Red dyes, are not approved for use as food
colorants and are considered likely carcinogens or genotoxins. So, a turmeric
raw material or product laced with any of these artificial color enhancers not
only represents a direct health challenge from the illegal colorants, it is
also not likely true turmeric and therefore would not convey any of the health
benefits expected from the real thing.
In addition, unlabeled
fillers or excipients, such as gluten-containing flours (e.g., wheat) or
allergen-containing materials (e.g., nuts) have been reported in turmeric
products.93 These represent a health hazard to those consumers with
sensitivities, allergies or other unfavorable reactions to such substances.
*Curcumin
is the common or trivial name given to the chemical compound diferuloylmethane,
or (1E,6E)-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione (see
compound 1 in Figure 1). The term
“curcumin” is also used in the dietary supplement industry to describe a
turmeric extract containing a natural ratio of curcumin, demethoxycurcumin, and
bisdemethoxycurcumin – the three morst abundant curcuminoides in C. longa. To avoid confusion, all
extracts made from C. longa, whether
these extracts are enriched in curcuminoids or not, will be indicated as
turmeric extracts in this document. Curcuminoids is a common or trivial name given to the overall class of diarylheptanes, which includes not only the curcumins (1-3), but any related monor compounds in C. longa and as yet undiscovered, related compounds of in other species of Curcuma. Curcuminoids is a common or trivial name given to the overall class of diarylheptanes, which includes not only the curcumins (1-3), but any related minor compounds in C. longa and as yet undiscovered, related compounds in other species of Curcuma.
†D.
Mundkinajeddu (Natural Remedies, Bangalore, India) email to S. Gafner, January
24, 2020.
‡Jiang huang refers to the rhizome derived from Curcuma
longa. The Pharmacopoeia of the
Peoples Republic of China lists the dry tuberous root of C. longa, C. kwangsiensis, C. phaeocaulis,
and C. wenyujin as yu jin. Curcuma longa is specified as huang
si yu jin.
§The
Plant List classifies C. ecalcarata
as a synonym of C. aurantiaca, while
both C. malabarica and C. raktakanta are considered synonyms of
C. zedoaria.
10. References
- Smith T, Gillespie M, Eckl
V, Knepper J, Reynolds CM. Herbal supplement sales in US increase by 9.4% in 2018.
HerbalGram 2019; 123:62-73.
- Bejar E. Adulteration
of Turmeric (Curcuma longa) Root and Rhizome, and Root and Rhizome Extracts. Austin, TX: ABC-AHP-NCNPR Botanical
Adulterants Prevention Program; Botanical Adulterants Prevention Bulletin. 2018; 1-11.
- Spices Board of India website. Spices Board India; 2017. http://www.indianspices.com/spice-catalog/turmeric-1.
Accessed May 8, 2019.
- Kays, SJ, Silva Dias JC. Common names of commercially cultivated vegetables of the
world in 15 languages. Econ Bot. 1995; 49:115-152.
- Nadkarni KM, Nadkarni AK. Dr.
K. M. Nadkarni's Indian Materia Medica: With Ayurvedic, Unani-tibbi, Siddha, Allopathic, Homeopathic,
Naturopathic & Home Remedies, App. & Indexes. 3rd ed.
Bombay, India: Popular Book Depot; Popular Prakashan; 1982.
- Biodiversity of India: a Wiki resource for Indian biodiversity.
2010. http://www.biodiversityofindia.org/index.php?title=Curcuma_longa.
Accessed May 22, 2018.
- Aggarwal BB. Health benefits of herbs and spices. How new
findings will disrupt global spice industry? Presented at: European Spice Association
General Assembly, May 2017; Bordeaux, France. https://www.esa-spices.org/download/health-benefits-of-herbs-and-spices-bharat-aggarwal.pdf.
Accessed May 9, 2019.
- Moley T, Foster S, Awang D, Hu SY, Kartesz JT, Tucker AO. Herbs of Commerce. 1st ed. Austin, TX:
American Herbal Products Association; 1992.
- McGuffin M, Kartesz JT, Leung AY, Tucker AO. Herbs of Commerce. 2nd ed. Silver
Spring, MD: American Herbal Products Association; 2000.
- Nguyen VD. Medicinal
Plants of Vietnam, Cambodia and Laos. Santa Ana, CA: Mekong Printing; 1993.
- Flora of China. Missouri Botanical Garden & Harvard
University Herbaria. http://www.efloras.org/florataxon.aspx?flora_id=2&taxon_id=200028370.
Accessed May 22, 2018.
- Zhao Z, Chen H. Yu jin.
Chinese Medicinal Identification. An Illustrated
Approach. Taos, NM: Paradigm Publications; 2014; 201.
- Mills S, Bone K. Principles
and Practice of Phytotherapy: Modern Herbal Medicine. Edinburgh: Churchill
Livingstone; 2000.
- Quisumbing EA. Medicinal
Plants of the Philippines. Quezon City, Philippines: Katha Publishing Co;
1978.
- Lim TK. Curcuma longa.
Edible Medicinal and Non-Medicinal
Plants, Vol. 12. Modified Stems, Roots, Bulbs. Cham, Switzerland; 2016;
241-362.
- Flowers of India. http://www.flowersofindia.net/catalog/slides/Turmeric.html.
Accessed May 22, 2018.
- Asolkar LV, Kakkar KK, Chakre OJ. Second Supplement to Glossary of Indian Medicinal Plants with Active
Principles, Part-I (A-K) (1965-1981). New Delhi, India: Publications &
Information Directorate, CSIR; 1992.
- The Plant List. Version 1.1. http://www.theplantlist.org. Accessed
May 22, 2018.
- Williamson E. Major
Herbs of Ayurveda. Churchill Livingstone, New York; 2002.
- Engels G. Turmeric. HerbalGram.
2009; 84:1-3.
- Li S, Yuan W, Deng G, Wang P, Yang P, Aggarwal B. Chemical
composition and product quality control of turmeric. Pharm Crops 2011;2:28-54.
- Priyadarsini KI. The chemistry of curcumin: from extraction
to therapeutic agent. Molecules 2014;19:20091-20112.
- Curcuma longa L.
Botanical Voucher Speciman. American Herbal Products Association website. http://www.botanicalauthentication.org/index.php/Curcuma_longa_(rhizome)#Botanical_Voucher_Specimen.
Accessed June 1, 2018.
- Ravindran PN, Nirmal Babu K, Shiva KN. Botany and crop
improvement of turmeric. In: Ravindran PV, Nirmal Babu K, Sivaraman K, eds. Turmeric: the Genus Curcuma. Boca Raton, FL: CRC Press; 2007;
15-70.
- Seema R. PhD Thesis:
Micromorphological Distinction of Indian Curcumas. Lucknow, India:
Department of Botany, University of Lucknow; 2015.
- Upton R, Graff A, Jolliffe G, Länger R, Williamson E, Swisher
D. Botanical Pharmacognosy—Microscopic
Characterization of Botanical Medicines. CRC Press, Taylor & Francis
Group, Boca Raton. 2011; 339-341.
- Eschrich W. Pulver-Atlas der Drogen. 7 ed.
Stuttgart, Germany: Deutscher Apotheker Verlag; 1999.
- Tandon N, Sharma P, Gupta AK, eds. Quality Standards of Indian Medicinal Plants. Vol 8. New Delhi,
India: Indian Council of Medical Research; 2010; 138-148.
- Tandon N, Sharma P, Gupta AK, eds. Quality Standards of Indian Medicinal Plants. Vol 6. New Delhi,
India: Indian Council of Medical Research; 2008; 101-109.
- Tandon N, Sharma P, Gupta AK, eds. Quality Standards of Indian Medicinal Plants. Vol 7. New Delhi,
India: Indian Council of Medical Research; 2008; 67-77.
- Amel B. Microscopic analysis of Curcuma longa using multivariate test. Int J Pharmacognosy. 2015; 2:173-177.
- Jan HU, Rabbani MA, Shinwari
ZK. Assessment of genetic
diversity of indigenous turmeric (Curcuma
longa L.) germplasm from Pakistan using RAPD markers. J Med Plants Res. 2011; 5:823-830.
- Ashraf K, Ahmad A, Shah SAA, Mujeeb M. Genetic diversity
in accessions of Indian turmeric (Curcuma longa L.) using RAPD markers. Int J Pharm Pharm Sci. 2017; 9:288-291.
- Verma S, Singh S, Sharma S, Tewari SK; Roy RK, Goel AK,
Rana TS. Assessment of genetic
diversity in indigenous turmeric (Curcuma
longa) germplasm from India using molecular markers. Physiol Mol Biol Plants. 2015,
21:233-242.
- Arya N, Prakash O, Kumar S, Vivekanand, Pant AK. Curcumin profiling and genetic diversity of
different accessions of Curcuma longa
L. Asian Pac J Trop Dis. 2016; 6:70-74.
- Singh S, Panda MK, Nayak S. Evaluation of genetic diversity in turmeric (Curcuma longa L.) using RAPD and ISSR markers. Ind Crops Products. 2012; 37:284-291.
- Syamkumar S, Sasikumar B. Molecular marker based genetic diversity
analysis of Curcuma species from
India. Sci Hort. 2007; 112:235-241.
- Angel GR, Makeshkumar T, Mohan
C, Vimala B, Nambisan B. Genetic diversity analysis of starchy Curcuma species
using RAPD markers. J Plant Biochem & Biotechnol. 2008;
17:173-176.
- Theanphong
O, Thanakijcharoenpath W, Palanuvej C, Ruangrungsi N, Rungsihirunrat K. RAPD
marker for determination of phylogenetic relationships of 15 Curcuma species from Thailand. Bull Health Sci Tech. 2016; 14:45-56.
- Minami M, Nishio K, Ajioka Y, Kyushima H, Shigeki K, Kinjo K, Yamada K, Nagai M, Satoh K, Sakurai Y. Identification of Curcuma
plants and curcumin content level by DNA polymorphisms in the trnS-trnfM
intergenic spacer in chloroplast DNA. J Nat Med. 2009; 63:75-79.
- Nahar L, Sarker SD. Phytochemistry of the genus Curcuma.
In: Turmeric: the Genus Curcuma,
Ravindran PN, Nirmal Babu K, Sivaraman K., Eds. CRC Press: Boca Raton, 2007; 71-106.
- Golding BT, Pombo E, Christopher JS. Turmerones:
isolation from turmeric and their structure determination. J Chem Soc Chem Commun. 1982; 6:363-364.
- Sharma RK, Misra BP, Sarma T, Bordoloi AK, Pathak MG,
Leclercq PA. Essential oils of Curcuma longa L. from Bhutan. J Essent Oil Res. 1997; 9:589-592.
- Imai S, Morikiyo M, Furihata K, Hayakawa Y, Seto H.
Turmeronol A and turmeronol B, new inhibitors of soybean lipoxygenase. Agric
Biol Chem. 1990; 54:2367-2371.
- Chen JJ, Tsai CS, Hwang TL, Shieh PC, Chen JF, Sung PJ.
Sesquiterpenes from the rhizome of Curcuma longa with inhibitory
activity on superoxide generation and elastase release by neutrophils. Food
Chem. 2010; 119:974-980.
- Gleason K, Shine JP, Shobnam N, Rokoff LB, Suchanda HS, Ibne Hasan MO, Mostofa
G, Amarasiriwardena C, Quamruzzaman Q, Rahman M, Kile ML, Bellinger DC, Christiani
DC, Wright RO, Mazumdar M. Contaminated turmeric is a potential
source of lead exposure for children in rural Bangladesh. J Environ Public Health. 2014; 2014:5.
- Cowell W, Ireland T, Vorhees D, Heiger-Bernays W. Ground
turmeric as a source of lead exposure in the United States. Public Health Reports. 2017;
132:289-293.
- Spices USA Inc. issues alert on elevated levels of lead
in ground turmeric. US Food and Drug Administration; 2016. https://www.fda.gov/Safety/Recalls/ucm523561.htm. Accessed June 28, 2018.
- Angelon-Gaetz
KA, Klaus C, Chaudhry EA, Bean K. Lead in spices, herbal remedies, and ceremonial powders sampled from home investigations for children with elevated blood lead levels — North Carolina, 2011-2018. Morbidity and Mortality Weekly Report. 2018; 67(46):1290–1294. doi:10.15585/mmwr.mm6746a2. https://www.cdc.gov/mmwr/volumes/67/wr/mm6746a2.htm?s_cid=mm6746a2_e.
Accessed January 25, 2019.
- Forsyth JE,
Nurunnahar S, Islam SS, Baker M, Yeasmin D, Islam MS, Rahman M, Fendorf S,
Ardoin NM, Winch PJ, Lubye
SP. Turmeric means “yellow” in Bengali: Lead chromate pigments added to
turmeric threaten public health across Bangladesh. Environ Res. 2019;
179:108722. doi:
10.1016/j.envres.2019.108722
- Forsyth JE, Weaver KL, Maher K, Islam MS, Raqib R, Rahman
M, Fendorf S, Luby SP. Sources of blood lead exposure in rural Bangladesh. Environ
Sci Technol. 2019; 53:11429-11436.
- Dhakal S, Chao K,
Schmidt W, Qin J, Kim M, Chan D. Evaluation of turmeric powder adulterated with Metanil Yellow using FT-Raman and FT-IR spectroscopy. Foods. 2016;5:36. doi:10.3390/foods5020036.
- Dixit S, Purshottam SK, Khanna SK, Das M. Surveillance of the
quality of turmeric powders from city markets of India on the basis of curcumin
content and the presence of extraneous colours. Food Addit Contam Part A. 2009; 26:1227-1231.
- Opinion of the Scientific Panel on Food
Additives, Flavourings, Processing Aids and Materials in Contact with Food. The
EFSA Journal. 2005; 263:1-71. https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/j.efsa.2005.263. Accessed February 10, 2019.
- Salmén R, Fjærtoft Pedersen B, Malterud KE. Sudanrot G als
Zusatz in Gelbwurzel (Curcuma longa
L.). Z Lebensm Unters
Forsch. 1987; 184:33-34.
- Nelson KM, Dahlin
JL, Bisson J, Graham J, Pauli GF, Walters
MA. The essential
medicinal chemistry of curcumin. J Med Chem. 2017; 60:1620-1637.
- Sen AR, Gupta PS, Dastidar NG. Detection of Curcuma zedoaria and Curcuma aromatica in Curcuma longa (turmeric) by thin layer
chromatography. Analyst. 1974; 99:153-155.
- Mezzasalma V,
Ganopoulos I, Galimberti A, Cornara L, Ferri E, Labra M. Poisonous or non-poisonous
plants? DNA-based tools and applications for accurate identification. Int J Leg Med. 2017; 131:1-19.
- Balakrishnan KV. Postharvest technology and processing of
turmeric. In: Ravindran PN, Nirmal Babu K, Sivaraman K, eds. Turmeric: the Genus Curcuma. Vol 45. Boca Raton, FL: CRC Press;
2007: 193-256.
- Booker A, Frommenwiler D, Johnston D, Umealajekwu C, Reich E,
Heinrich M. Chemical variability along the value chains of turmeric (Curcuma longa): A comparison of nuclear
magnetic resonance spectroscopy and high performance thin layer chromatography.
J Ethnopharmacol. 2014;152:292-301.
- Sasikumar B, Syamkumar S, Remya R, Zachariah TJ. PCR based
detection of adulteration in the market samples of turmeric powder. Food Biotechnol. 2004;18:299-306.
- Parvathy VA, Swetha VP, Sheeja TE, Sasikumar B. Detection of
plant-based adulterants in turmeric powder using DNA barcoding. Pharm Biol. 2015;53:1774-1779.
- Dhanya K, Sasikumar B. Molecular marker based adulteration
detection in traded food and agricultural commodities of plant origin with
special reference to spices. Curr Trends
Biotechnol Pharm. 2010;4:454-489.
- Dhanya K, Syamkumar S, Siju S, Sasikumar B. Sequence
characterized amplified region markers: A reliable tool for adulterant
detection in turmeric powder. Food Res Int. 2011;44:2889-2895.
- Staesche K,
Schleinitz H. Curcuma. In: Hänsel R, Keller K, Rimpler H, Schneider G, eds. Hagers Handbuch der Pharmazeutischen Praxis,
Band 5. Drogen A-D. Berlin & Heidelberg, Germany: Springer-Verlag; 1992:1084-1102.
- Tonnesen HH, Karlsen J,Grislingaas A-L, Balakrishnan KVN, Ayyappan P, Verghese J. Studies on curcumin and curcuminoids. XXI. Variation in the content of
curcuminoids in Curcuma longa L. and Curcuma aromatica Salisb.
from India during one season. Z Lebensm Unters Forsch. 1992;194:570-572. https://link.springer.com/article/10.1007%2FBF01185486. Accessed May 17, 2019.
- Akter J, Hossain MA, Hossain MA, Takara K,
Islam MZ, Hou DX. Antioxidant
activity of different species and varieties of turmeric (Curcuma spp.) against Fusarium solari sensu lato. Pharm Chem
J. 2018;52:320-325.
- HPTLC Association: Turmeric rhizome (Curcuma longa).
HPTLC Association. http://www.hptlc-association.org/methods.cfm. Accessed May 17, 2019.
- Dosoky NS, Setzer WN. Chemical composition and biological
activities of essential oils of Curcuma species. Nutrients. 2018;10(9): pii: E1196. doi: 10.3390/nu10091196.
- Blaschek W, Frohne D, Loew D. Curcumae xanthorrhizae
rhizoma. In: Blaschek W, ed. Wichtl
-Teedrogen und Phytopharmaka. Stuttgart, Germany: Wissenschaftliche
Verlagsgesellschaft mbH; 2016: 213-215.
- Avula B, Wang YH,
Khan IA. Quantitative
determination of curcuminoids from the roots of Curcuma longa, Curcuma
species and dietary supplements using an UPLC-UV-MS method. J Chromatograph Separat Techniq. 2012;
3:120-125.
- Tohda C, Nakayama N, Hatanaka F, Komatsu K. Comparison of
anti-inflammatory activities of six Curcuma
rhizomes: a possible curcumin-independent pathway mediated by Curcuma phaeocaulis extract. eCAM. 2006; 3:255-260.
- Tiwari M, Agrawal R, Pathak AK, Rai AK, Rai GK. Laser-induced breakdown
spectroscopy: an approach to detect adulteration in turmeric. Spectroscopy Lett. 2013; 46:155-159.
- Food Safety and Standards Authority of India (FSSAI). Manual of simple methods for testing
of common adulterants in food.
New Delhi, India. 2017: 1-55. See also: https://www.turmericforhealth.com/general-info/how-to-test-turmeric-powder-for-quality-and-avoid-adulterated-products. Accessed August 15, 2019.
- Feng F, Zhao Y, Yong W,
Sun L, Jiang G, Chu X. Highly sensitive and accurate screening of 40 dyes in
soft drinks by liquid chromatography–electrospray tandem mass spectrometry. J Chromatogr B. 2011; 879:1813-1818.
- Dhakal S, Schmidt WF, Kim
M, Tang X, Peng Y, Chao K. Detection of additives and chemical contaminants in
turmeric powder using FT-IR spectroscopy. Foods. 2019; 8:143-157.
- Dixit S, Khanna SK, Das M. A simple 2-directional high-performance
thin-layer chromatographic method for the simultaneous determination of
curcumin, metanil yellow, and sudan dyes in turmeric, chili, and curry powders. J AOAC Int. 2008; 91:1387-1396.
- US Pharmacopeia, Powdered Turmeric. USP 42-NF 37. Rockville, MD: United States Pharmacopeial
Convention; 2019:5257.
- US Pharmacopeia, Powdered
Turmeric Extract. USP 42-NF 37.
Rockville, MD: United States Pharmacopeial Convention; 2019:5258.
- US Pharmacopeia, Curcuminoids.
USP 42-NF 37. Rockville, MD: United
States Pharmacopeial Convention; 2019:4872.
- European Pharmacopoeia,
Turmeric Rhizome: Curcumae longae rhizoma. 01/2015:2543.
- European Pharmacopoeia,
Javanese Turmeric Rhizome: Curcumae zanthorrhizae rhizoma. 01/2015:1441.
- ISO 5566.
Turmeric—Determination of colouring power—Spectrophotometric method. 1982.
- ASTA
Analytical Method 18.0. Curcuminoids content of turmeric spice and oleoresins.
Latest revision: April 2019.
- Windarsih
A, Rohman A, Swasono RT. Application of H-NMR metabolite fingerprinting
and chemometrics for the authentication of Curcuma longa adulterated
with Curcuma manga. J App Pharm Sci. 2018; 8:75-81.
- Windarsih A, Rohman A, Swasono RT. Authentication of
turmeric using proton-nuclear magnetic resonance spectroscopy and multivariate
analysis. Int J App Pharm. 2018; 10:174-180. DOI:
http://dx.doi.org/10.22159/ijap.2018v10i6.29014.
- Mudge E, Chan M,
Venkataraman S, Brown PN. Curcuminoids in turmeric roots and supplements:
method optimization and validation. Food
Anal Methods. 2016;9:1428-1435.
- Mudge E, Brown PN. Determination of curcuminoids in turmeric raw
materials and dietary supplements by HPLC: Single-Laboratory Validation, First
Action 2016.16. J AOAC Int. 2018;101:203-207.
- Wichitnithad W, Jongaroonngamsang
N, Pummangura S, Rojsitthisak P. A simple isocratic HPLC method for the
simultaneous determination of curcuminoids in commercial turmeric extracts. Phytochem Anal. 2009;20:314-319.
- Paramapojn S, Gritsanapan
W. Free radical scavenging activity determination and quantitative analysis of
curcuminoids in Curcuma zedoaria rhizome extracts by HPLC method Curr Sci. 2009;97:1069-1073.
- Guddadarangavvanahally KJ,
Lingamullu JMR, Kunnumpurath KS. Improved HPLC method for the determination of
curcumin, demthoxycurcumin, and bisdemethoxycurcumin. J Agric Food Chem. 2002;50:3668-3672.
- Ferreira LA, Henriques OB,
Andreoni AA, Vital GR, Campos MM, Habermehl GG,
de Moraes VL. Antivenom and biological effects of ar-turmerone
isolated from Curcuma longa (Zingiberaceae).
Toxicon. 1992;30:1211-1218.
- Parvathy
VA, Swetha VP, Sheeja TE, Sasikumar B. Detection of plant-based adulterants in
turmeric powder using DNA barcoding. Pharm
Biol. 2015;53:1774-1779.
- Barbosa C,
Nogueira S, Gadanho M, Chaves S. Study on commercial spice and herb products
sing Next-Generation Sequencing (NGS). J AOAC Int. March 1, 2019; 102(2):369-375.
- Beta Analytic Testing Laboratory: https://www.betalabservices.com/curcumin-adulteration/.
Accessed August 22, 2019.
|