Issue:
97
Page: 58-67
Enhancing Quality Control of Botanical Medicine in the 21st Century from the Perspective of Industry:
The use of chemical profiling and DNA barcoding to ensure accurate identity
by Yuan-Chun Ma, PhD, Shi-Lin Chen, PhD, Michelle E. Thibault, PhD, Jie Ma
HerbalGram. 2013; American Botanical Council
Introduction
Herbal products are growing increasingly popular in North
America, including those derived from North American and European herbal
traditions, Traditional Chinese Medicine, and Ayurveda. However, there are
problems with many products on the market today. Misidentification of plant
species, adulteration with counterfeit ingredients, insufficient quantities of
the known primary active ingredients, and spiking with marker compounds
commonly occur.
Manufacturers of many
consumer products that include medicinal plant ingredients have an obligation
to ensure that the products they sell are genuine and safe; marketers of food
products containing so-called “medicinal botanicals,” including dietary
supplements in the United States, usually have no regulatory requirements to
ensure that their products are effective (unless certain limited health-related
claims are made).
Adulteration via
species substitution may occur accidentally or intentionally using closely
related or completely unrelated species. Thus, the first step in quality
control must be proper identification of each ingredient. Botanical medicinal
materials are identified by their organoleptic (color, taste, fragrance, etc.),
morphological (shape), microscopic, and/or chemical chromatographic characteristics,
e.g., by the use of thin-layer
chromatography (TLC) and/or other chromatographic methods. Someone who is not
sufficiently knowledgeable of the plants in question will not be able to
accurately identify botanical ingredients. Many closely related species share
morphological features and/or common names, which can lead to potential
confusion and accidental adulteration. Furthermore, most herbs are sold
partially processed — dried, cut into pieces, shredded, or even powdered — such
that macroscopic morphological identification of the plant part (flowers,
leaves, roots, etc.) is no longer possible, although microscopic and
chromatographic identification can still be performed.*
Reliable analytical
methods are needed to supplement these typical protocols for identification of
botanical medicinal materials. Chemical profiling using TLC, high-performance
TLC (HPTLC), gas chromatography (GC), and high-performance liquid
chromatography (HPLC) is common, and such profiles are documented in herbal
monographs found in resources such as the American
Herbal Pharmacopoeia, the United
States Pharmacopeia, the Pharmacopoeia
of the People’s Republic of China, and the Journal of the Association of Official Analytical Chemists. In
addition, techniques such as near-infrared (NIR) and nuclear magnetic resonance
(NMR) spectroscopy are becoming more common in the scientific community.
However, it must be considered that the chemical profile of an herb may vary
due to factors such as growth stage, plant part, geography, and post-harvest
processing and storage, which is why multiple reference materials must be used
to statistically overcome such variations.†
DNA barcoding is
growing in popularity as a means of species identification.1 In
October 2011, the US Food and Drug Administration (FDA) formally approved the
use of DNA barcoding for the identification of seafood in order to counteract
the widespread practice of substituting and mislabeling cheaper or undesirable
species of fish and seafood as more expensive species.2 Simultaneous
with this announcement, FDA released a validated laboratory method for the DNA
barcoding of fish species for the purposes of regulatory compliance.3
We propose that DNA barcoding be added in the future to the quality control
toolbox for medicinal botanical identification, alongside organoleptic,
microscopic, and chemical profiling.
What Is DNA Barcoding?
DNA barcoding is the
use of a short region of DNA to identify species.4 The first step to
obtaining a DNA barcode is the extraction of DNA from a small sample of the
specimen. Second, the selected barcode region undergoes polymerase chain
reaction (PCR) amplification, or copying. Third, the PCR-amplified product is
purified and sequenced (the order of nucleotides read). Finally, the DNA
sequence is compared to the sequences in a library to identify the species in
question. Figure 1 illustrates the DNA barcoding process.

PCR amplification
entails multiple cycles of a three-phase process. The double-stranded DNA is
denatured (separated into its individual strands) at a high temperature. Next,
the temperature is lowered and sequence-specific primers (short sequences of 20
or so nucleotides) attach to sites neighboring the target sequence. Primers are
required as the DNA polymerase can only add new nucleotides to an existing
piece of double-stranded DNA. Finally, the DNA polymerase uses the single
strand of DNA as a template to extend the sequence from the primers. This new
product then becomes the template for the next cycle. The cycles are repeated 20
to 30 times, generating thousands to millions of copies of the target DNA
sequence.
Standardization,
minimalism, and scalability are key factors in the application of DNA
barcoding. Practically speaking, this means that one or a few standard regions
of a limited number of DNA base pairs (usually 200 to 1,000) must be chosen so
that they can be sequenced readily in a large and varied sample set, enabling
comparison of the data and allowing for species identification.5 As
a corollary, the inter-species variation in the DNA sequence should be much
larger than the intra-species variation. In animals, a fragment of the
cytochrome c oxidase 1 (CO1) gene has
been accepted as the standard DNA barcode. In plants, however, no single region
has been found that meets all of the criteria of universality (ease of
sequencing in all land plants), sequence quality, and species discrimination.
The Consortium for the Barcode of Life (CBOL) has proposed the combination of
the matK and rbcL genes as the core plant barcode, though it recognizes that matK + rbcL may at times need to be supplemented with other markers.6
Specifically, matK cannot always be
amplified and sequenced, though its species discrimination is high, while rbcL is easy to amplify and sequence,
but its species discrimination is low. The internal transcribed spacers of
nuclear ribosomal DNA (nr ITS/ITS2) and the chloroplast intergenic spacer psbA-trnH have been proposed as
alternates to matK + rbcL,5 and, in fact, the
China Plant Barcode of Life group has suggested the addition of ITS, or ITS2
when ITS cannot be successfully sequenced, to the core plant barcode of matK + rbcL.7
 The biggest challenge
thus far in DNA barcoding of plants has been that good, universal primers for
plant marker barcodes can be difficult to design. Amplification and/or
sequencing of a given marker may be possible only in certain families of
plants. For a particular marker, genetic gaps between species may be large in
some groups of plants, but not in others.8 For these reasons, it
appears that several markers, alone or in combination, will be required for the
DNA barcoding of plants, rather than the single CO1 marker prevalent in the DNA
barcode analysis of animals.
A further problem
with DNA barcoding of plants is that many plants lack barcodes altogether, and
there is not yet a universal database of plant barcodes.9 However,
as a major use of DNA barcoding is the identification of unknown specimens,
non-chemistry specialists such as customs officers, producers of traditional
medicines, pharmaceutical manufacturers, and forensics investigators may
welcome a relatively rapid and simple — albeit still imperfect — method for the
identification of botanical products.10 Nevertheless, a great deal
of work is still needed before DNA barcoding of plants can be considered
sufficiently reliable for widespread practical application.
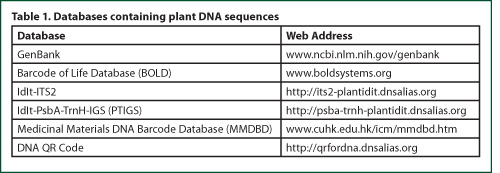
GenBank is a database
of all publicly available DNA sequences and is part of the International
Nucleotide Sequence Database Collaboration, which also includes the DNA DataBank
of Japan and the European Molecular Biology Laboratory. These three
organizations exchange data on a daily basis. Databases specific to DNA
barcodes include the Barcode of Life Database,11 which is based on
the matK + rbcL combination, as well as the IdIt-ITS2 and PTIGS (IdIt-psbA-trnH-IGS) databases, which are
based on ITS2 and psbA-trnH,
respectively.12,13
Of particular
interest and use to those in industries or markets that utilize medicinal
plants as ingredients is the Medicinal Materials DNA Barcode Database (MMDBD).14
At the time of its 2010 publication, the database contained more than 18,000
sequences from 1,259 species, representing 66.5% and 84.5% of the medicinal
materials listed in the 2005 Pharmacopoeia
of the People’s Republic of China and the American Herbal Pharmacopoeia, respectively. As of May 2012, the
MMDBD featured more than 31,000 barcode sequences from more than 1,650 indexed
species. Core and supplementary DNA barcodes for medicinal materials listed in
the above pharmacopeias and other sources are included, as well as information
on adulterants and substitutes, photographs of the medicinal materials, PCR
conditions, and literature references. The database can be searched by keyword
or sequence similarity, and researchers can upload their DNA barcode sequences
to help expand the database.‡
Finally, Liu et al. have established a web
application that will convert a DNA barcode into a two-dimensional Quick
Response (QR) Code for use in practical applications — in essence, barcoding
the barcode.15 The user can retrieve the DNA sequence and QR code
for a species of interest, convert a sequence to a QR code and vice-versa, or
search the database using a QR code to identify a sample. This leads one to
envision a system in which an herbal material is labelled with its QR DNA
barcode as a means of inventory tracking.
Undoubtedly, DNA
barcoding of plants will improve with advances in PCR amplification and DNA
sequencing technology. Identification of plants will be enhanced with better
access to authenticated botanical DNA libraries that contain more species and
more samples of each species.
DNA Barcoding of Botanical Medicines
According to surveys
in China, medicinal plants comprise more than 11,000 species in 2,300 genera
and nearly 400 families. Quick and accurate authentication of these plants and
their adulterants can be difficult on an international trade scale. Shi-Lin
Chen, PhD, an author of this article,
and colleagues at the Institute of Medicinal Plant Development in
Beijing have been dominant in the field of DNA barcode analysis of botanical
medicines. Chen et al. investigated
different DNA regions for the purpose of barcoding plants found in the
traditional Chinese Materia Medica,
both in terms of PCR efficiency and species identification.10 The
PCR efficiency for both ITS2 and psbA-trnH
was greater than 90 percent. Furthermore, psbA-trnH
was more successful for some plants such as ferns. The identification rate of
ITS2 was 92.7% and 99.8% at the species and genus levels, respectively, for
6,685 samples from 4,800 species in 753 genera of 193 families. In contrast, psbA-trnH correctly identified only
about 70 percent of the species, though it was more than 95 percent accurate at
the genus level for 2,108 samples from 1,433 species in 551 genera of 135
families. They proposed the use of ITS2, supplemented by psbA-trnH, as the standard barcode for international trade and safe
use of medicinal plants.
In an additional
study, Yao et al. evaluated the ITS2
sequences of 50,790 plant samples available in GenBank. Species identification
rates ranged from 67 percent to 88 percent.12 A recent review
article by Chen et al. summarized
their work on the families Rosaceae, Fabaceae, Asteraceae, Rutaceae,
Euphorbiaceae, Polygonaceae, and the genera Paris
(Melanthiaceae), Lonicera
(Caprifoliaceae), Dendrobium
(Orchidaceae), Cistanche
(Orobanchaceae), Panax (Araliaceae),
and Datura (Solanaceae), as well as
medicinal pteridophytes and cortex herbs (medicinal materials from the bark of
stems or roots).16
The Journal of Systematics and Evolution
recently published a special issue on plant DNA barcoding in China.17
In particular, Li et al. reviewed
more than 125 studies on the application of DNA barcodes to the identification
of more than 75 different Chinese herbal medicinal materials.18 They
concluded that DNA barcoding of medicinal plants is still a work in progress,
but that it holds great promise for future applications in taxonomy,
biodiversity, conservation, the pharmaceutical industry, and forensics; the authors
proposed that future work should focus on reliable species identification and
barcoding multiple samples of each species to help build the reference database
for Chinese medicinal plants. The Chinese Pharmacopoeia Commission, recognizing
the value of DNA barcoding for the authentication of medicinal materials, has
included protocols and DNA barcodes for some animal-derived traditional Chinese
medicines in the 2010 edition of the Pharmacopoeia
of the People’s Republic of China, such as Wushaoshe (Chinese rat snake; Zaocys
dhumnades, Colubridae) and Qishe
(Chinese moccasin; Agkistrodon acutus,
Viperidae).18,19 Work is underway on drafting guidelines for the
identification of Chinese herbal medicines using DNA barcodes, potentially to
be included in the 2015 edition (Hui Yao email to M. Thibault, September 5,
2012).
An Explanation of Chemical Profiling
DNA barcoding is an
excellent solution for identifying raw or dried plant products. However, many
botanical products are sold as liquid or powder extracts. The alcohol and heat
used during the extraction process filters out or eliminates most cellular data
and denatures proteins and DNA, rendering DNA barcoding unfeasible.
Consequently, chemical identification of marker compounds must be utilized.
Raw herbs and
extracts possess a characteristic botanical profile of phytochemicals.
Initially, one or two of these phytochemicals were used as marker compounds for
the purpose of qualitative and quantitative quality control, which led to
spiking with low-quality or fraudulent botanical extracts containing the marker
compounds by unscrupulous producers. With the technological advances of the
last 20 years, simultaneous analysis for multiple chemical constituents is
possible. Thus, many herbs and botanical extracts are now analyzed for several
marker compounds as a means of circumventing potential spiking issues. For
example, Rhodiola rosea
(Crassulaceae) root extracts were formerly standardized only for salidroside.
After the discovery of widespread substitution of other Rhodiola species for R. rosea,
the latter extracts are now standardized for salidroside and rosavins. Rosavins
are unique to R. rosea, whereas
salidroside is found across the Rhodiola
genus and in some plants outside the genus.20
HPTLC is a simple,
rapid, economical, and qualitative method of identification. It allows for the
natural variability within a plant and can be used even when many chemical
components of the sample are unknown. Reference compounds, plant samples, and
adulterants can be compared in a parallel, high-throughput fashion. In
addition, the multiple chemical components of an herb are often present in a
consistent ratio to one another. HPLC commonly is used to separate and quantify
these constituents, which results in a characteristic profile, or fingerprint,
of the herb or extract. Manufacturers can use these profiles to help optimize
their extraction procedures, such that the resultant extract has the same
profile as the initial raw herb. This is beneficial to herbalists, naturopaths,
integrative physicians, and other traditional medicine practitioners who have a
holistic view of herbs and healing.
Over the last 20
years, there have been thousands of publications discussing the HPLC profile of
popular herbs. As mentioned earlier, many pharmacopoeias include HPLC methods
and profiles for quality control in the botanical industry. The Canadian
Phytopharmaceuticals Corporation has established a proprietary database of HPLC
profiles for more than 100 North American, South American, European, Ayurvedic,
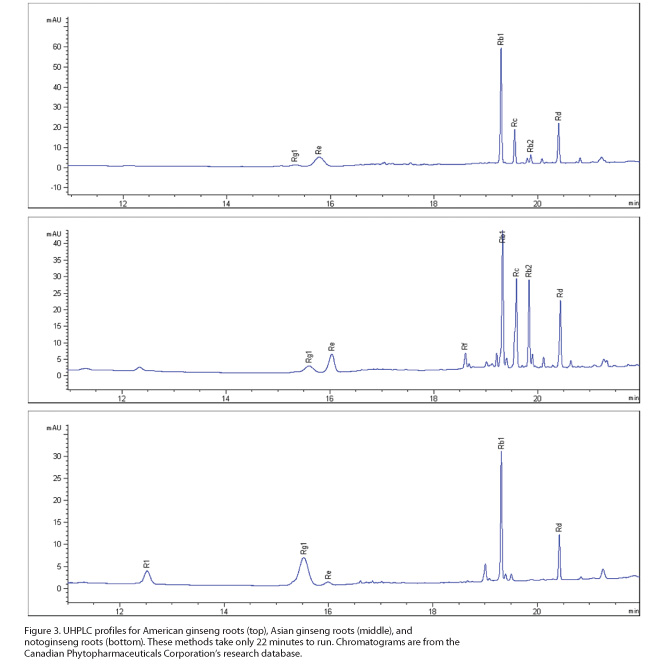
and traditional Chinese botanicals and extracts. Shown in Figure 2 are the HPLC
profiles developed by this HerbalGram
article’s co-author, Ma, and colleagues in the 1990s for American ginseng (Panax quinquefolius, Araliaceae), Asian
ginseng (P. ginseng), and notoginseng
(P. notoginseng).21,22
Each of these species has a characteristic ratio of ginsenosides that
distinguish one from the other.

With advances in
technology — such as the development of Ultra High-Performance Liquid
Chromatography (UHPLC, also commonly referred to as UPLC), gradient elution,
multi-wavelength detectors, and other types of detectors — analytical methods
have become much more powerful and simple. UHPLC offers significant time and
cost savings over conventional HPLC, due to its shorter run times and
concomitant reduced solvent usage. Thus, returning to the example of the three Panax species, the UHPLC profiles
developed in the 2010s are completed in half the time but maintain the same
appearance as the earlier HPLC profiles (Figure 3). In a further development, a
method recently was established in which the three Panax species, alone or in combination with Epimedium leaves (Berberidaceae), could be quantified in just four
minutes as compared to the 45 minutes required by the HPLC method.23
 Remedies developed by
traditional Chinese and other herbal medicine practitioners often involve
combinations of herbs. Method development for the HPLC fingerprinting of
formulated or combination products represents a breakthrough in the quality
control of botanical products. Individual herbs may have been analyzed by
different methods, using different HPLC columns, solvent gradients, or
detection wavelengths. Their profiles may overlap; hence, new methods must be
developed that will distinguish the profile for each herb, yet still allow for
analysis within a reasonable timeframe. The complexity of this task necessarily
increases with the number of herbs present in the combination product.
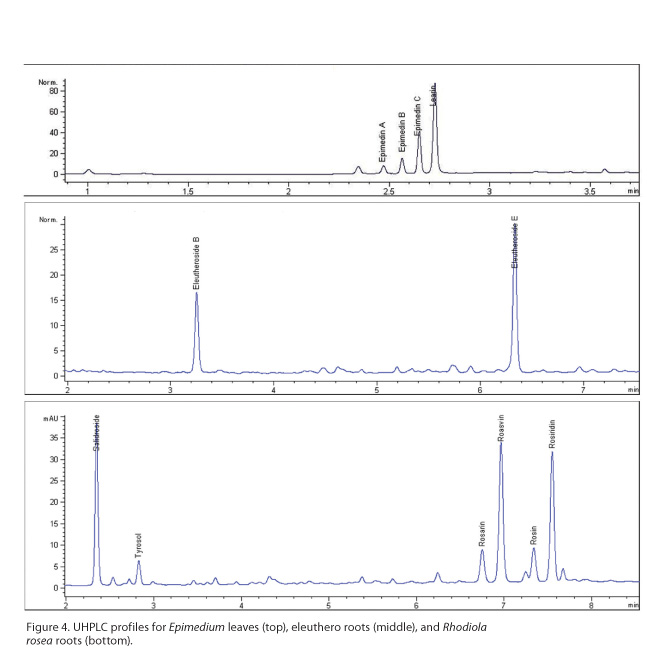
As an example,
consider a formulated product consisting of American ginseng roots, Epimedium koreanum (Berberidaceae)
leaves, eleuthero (Eleutherococcus
senticosus, Araliaceae) rhizomes, and R.
rosea roots. Such a combination may be used as a Western-style “Energy
Formula.” UHPLC profiles for the latter three herbs are shown in Figure 4, with
relevant marker compounds labeled. Run times range from four minutes for Epimedium to eight minutes for R. rosea24 and eleuthero.25
The UHPLC profile for the combination product (Figure 5), while complicated,
clearly shows the unique fingerprint of each herb, and the quantification of
more than 20 compounds is complete in only 22 minutes in a single run.
 Shuang-Huang-Lian (SHL) is a traditional Chinese formula comprised of Flos Lonicerae (Japanese honeysuckle; Lonicera japonica, Caprifoliaceae), Radix Scutellariae (Chinese skullcap; Scutellaria baicalensis, Lamiaceae), and
Fructus Forsythiae (forsythia; Forsythia suspensa, Oleaceae). It is
used commonly to treat upper respiratory illnesses. Ma et al. developed a UHPLC profile for SHL that is complete in seven
minutes (Figure 6),26 and extended the study to an “East-meets-West”
SHL-Echinacea combination (E. angustifolia and E. purpurea, Asteraceae).27
Technological and
analytical methodology development makes possible the qualitative and
quantitative analysis of multiple marker compounds in formulated products,
guaranteeing the quality of these products. Very few manufacturers currently
analyze combination products. Those that do are in a position to be leaders in
the marketplace.
Conclusion
The current industry
standards can and will change and improve according to market demands. Industry
must take the lead and set the benchmark for the quality control of botanical
extracts and Traditional Chinese Medicine, to counteract the erroneous belief
that herbal medicines are unregulated, untested, and ineffective. Combining the
applicable, reliable, and practical complementary techniques of DNA barcoding
and chemical profiling for the quality control of herbal products — from raw herb
to extract to finished product — will assure the delivery of high-quality,
safe, and efficacious products to market. Since DNA barcoding is not yet ready
for widespread implementation, an interim solution would be for botanical
product manufacturers to establish specifications that require testing and
conformance of the raw herbal material or extract with a pharmacopoeial
monograph. This includes organoleptic, microscopic, and chemical (TLC or HPLC)
profiling. Furthermore, by better validating the quality of botanical
ingredients used in products that may undergo robust pharmacological or
clinical studies, there should be a higher level of confidence and scientific
credibility in the clinical results.
Yuan-Chun Ma, PhD, founder, president, and CEO of Canadian
Phytopharmaceuticals Corporation (CPC) in Vancouver, BC, Canada, received his
doctorate in Pharmaceutical Sciences from the School of Pharmacy and Biomedical
Sciences at the University of Portsmouth in England. Author of over 60 research
and scientific publications, Dr. Ma is a guest professor with both the Chinese
Academy of Medical Sciences in Beijing and the Tongji Medical School, Huazhong
University of Science and Technology in Wuhan, China. He can be contacted at info@canphyto.com.
Professor Shi-Lin Chen, PhD, is director of the Institute of
Medicinal Plant Development, affiliated with the Chinese Academy of Medical
Sciences and Peking Union Medical College in Beijing. He obtained his PhD from
Chengdu University of Traditional Chinese Medicine and has been a Visiting
Professor at Hong Kong Polytechnic University. He holds a concurrent position
as the editor of such reputable Chinese medicinal research journals as China Journal of Chinese Materia
Medica, Chinese Medicine, Journal of Chinese Pharmaceutical
Science, Chinese Traditional and
Herbal Drugs, and World Science and
Technology. He has published more than
160 scientific papers.
Michelle E. Thibault, PhD, is a quality administrator at
CPC. She received her doctorate in Chemistry from the University of Guelph in
Canada, and is co-author of 15 papers and one patent.
Jie Ma is a PhD
candidate in the School of Pharmacy and Biomedical Sciences at the University
of Portsmouth and quality control manager at CPC.
* Responsible
companies should require their qualified suppliers to carry out suitable
identification tests, including appropriate macroscopic examination, prior to
particle size reduction and also require their supplier’s quality control unit
to retain samples of the whole, uncut starting material so that the companies
can trace back and re-test such material at a future date in the event of a
problem requiring rapid investigation, for example, a quality problem
(adulteration, contamination, etc.) or, in a worst-case scenario, a product
recall.
†This may seem
self-evident. In the context of pharmacopoeias mentioned in the previous
sentence, these factors are accounted for in the establishment of a monograph.
If a material does not test in conformance with all of the qualitative and
quantitative limits in the monograph, then a company producing botanical
products with the intention that they should provide a therapeutic or other
health benefit likely should reject the material as it would be indicative of one
or more problems at source, such as having been harvested at the wrong growing
stage (e.g., immature) or wrong plant
part (e.g., should be all leaf but
contains a high percentage of stems), or having been grown in the wrong climate
such that secondary metabolites never developed due to lack of stress
conditions.
‡The PPRC 2010
has 2,165 botanical monographs including Chinese Materia Medica crude drugs, crude drug preparations, prepared
slices, patent Chinese traditional medicines, oils and extracts. So far the AHP
has published 33 monographs.
References
- Cimino M. Ensuring the specific identity and quality of herbal products by the power of DNA. HerbalGram. 2010;86:50-57.
- DNA-based seafood identification. US Food and Drug Administration website. Available at: www.fda.gov/Food/FoodSafety/Product-SpecificInformation/Seafood/DNAspeciation/default.htm. Accessed July 6, 2012.
- Single laboratory validated method for DNA-barcoding for the species identification of fish for FDA regulatory compliance. Available at: www.fda.gov/Food/ScienceResearch/LaboratoryMethods/ucm237391.htm. Accessed July 6, 2012.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R SocLond B. 2003;270:313-321.
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6(5):e19254.
- CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106(31):12794-12797.
- China Plant BOL Group. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA. 2011;108(49):19641-19646.
- Li DZ, Liu JQ, Chen ZD, et al. Plant DNA barcoding in China. J Syst Evol. 2011;49(3):165-168.
- Hollingsworth PM. Refining the DNA barcode for land plants. Proc Natl Acad Sci USA. 2011;108(49):19451-19452.
- Chen SL, Yao H, Han JP, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5(1):e8613.
- Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol Ecol Notes. 2007;7:355-364.
- Yao H, Song JY, Liu C, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010;5(10):e13102.
- Liu C, Liang D, Gao T, et al. PTIGS-IdIt, a system for species identification by DNA sequences of the psbA-trnHintergenic spacer region. BMC Bioinformatics. 2011;12(Suppl 13):S4.
- Lou SK, Wong KL, Li M, But PPH, Tsui SKW, Shaw PC. An integrated web medicinal materials DNA database: MMDBD (Medicinal Materials DNA Barcode Database). BMC Genomics. 2010;11:402.
- Liu C, Shi LC, Xu XL, et al. DNA barcode goes two dimensions: DNA QR code web server. PLoS One. 2012;7(5):e35146.
- Chen SL, Pang XH, Yao H, Han JP, Luo K. Identification system and perspective for DNA barcoding Traditional Chinese Materia Medica. World Science and Technology – Modernization of Traditional Chinese Medicine. 2011;(5):747-754 and references therein.
- Special Issue: Plant DNA barcoding in China. J Syst Evol. 2011;49(3):165-283.
- Li M, Cao H, But PPH, Shaw PC. Identification of herbal medicinal materials using DNA barcodes. J Syst Evol. 2011;49(3):271-283.
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China. Beijing: Chemical Industry Press; 2010.
- Brown RP, Gerbarg PL, Ramazanov Z. Rhodiola rosea. A phytomedicinal overview. HerbalGram. 2002;56:40-52.
- Ma YC, Zhu J, Benkrima L, et al. A comparative evaluation of ginsenosides in commercial ginseng products and tissue culture samples using HPLC. J Herbs Spices Med Plants. 1996;3(4):41-50.
- Ma YC, Luo M, Malley L, Doucet M. Distribution and proportion of major ginsenosides and quality control of ginseng products. Chinese Journal of Medicinal Chemistry. 1996;6(1):11-21.
- Ma J, Ma YC, Wang D, et al. Simultaneous quantification of Panax and Epimedium species using Rapid Resolution Liquid Chromatography (RRLC). Nat Prod Commun. 2011;6(5):581-586.
- Ma YC, Wang XQ, Hou FF, et al. Rapid resolution liquid chromatography (RRLC) analysis for quality control of Rhodiola rosea roots and commercial standardized products. Nat Prod Commun. 2011:6(5):645-650.
- Ma YC, Wang XQ, Hou FF, et al. Simultaneous quantification of polyherbal formulations containing Rhodiola rosea L. and Eleutherococcus senticosus Maxim. using rapid resolution liquid chromatography (RRLC). J Pharm Biomed Anal. 2011;55:908-915.
- Ma YC, Wang XQ, Hou FF, et al. Rapid resolution liquid chromatography (RRLC) analysis and studies on the stability of Shuang-Huang-Lian preparations. J Pharm Biomed Anal. 2011;54:265-272.
- Ma J, Ma YC, Cai C, et al. Simultaneous quantification of Echinacea species, Flos Lonicerae, Radix Scutellaria and Fructus Forsythiae combinations by rapid resolution liquid chromatography. Nat Prod Commun. 2011;6(5):639-643.
|